Overview
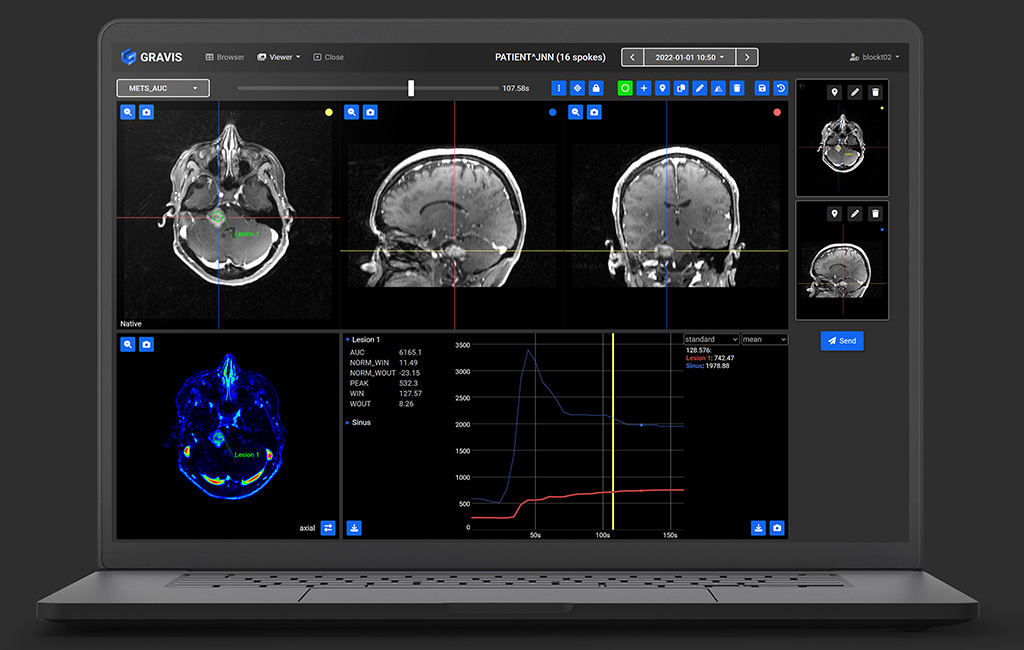
GRAVIS is a web-based open-source visualization and annotation tool designed for use with large, four-dimensional DICOM volumes. It was designed to display GRASP datasets reconstructed at high time resolutions without undue latency or processing delays.
Why use GRAVIS?
GRAVIS is designed for a small team that wants to view and annotate 4D GRASP datasets that are too bulky for their existing software tools. It may also be a useful starting point for development for related use-cases.
It currently does not implement a standard DICOM receiver. It can receive datasets from with both YarraServer and mercure using their respective SFTP transfer functionality. The former means that reconstruction outputs can flow from YarraServer directly to GRAVIS for viewing.
Alternatively, GRAVIS users can load datasets in on an ad-hoc basis if the files are sufficiently organized and already available on the server itself. At the moment, receiving DICOMs via the DICOM protocol relies on setting up and configuring mercure.
Features
- Navigate 4D dicom volumes in space and time without loading the entire dataset at once
- Annotate volumes with points and elliptical regions of interest
- Calculate summary statistics and time curves
- Multiple readers can collaborate on annotating a set of studies
- Store and transmit findings as DICOM Secondary Capture images
- Web-based viewer
Current limitations
- No support for standard DICOM transfers
- No fine-grained permissions or assigning studies to projects. Designed with small groups in mind.
- Not a fully-featured web imaging platform; you probably want OHIF for that.
- 2D regions of interest only, no freehand / polygonal ROIs.
- Regions of interest are limited to axial/sagittal/coronal planes
- Pre-processing pipelines are not configurable without making changes in the code. The code is designed to be fairly modular and easy to extend or modify, but it still requires accessing the backend to implement your own pipeline.
- Does not support non-isotropic images(?)
Workflow
GRAVIS uses a batch processing data flow. For each dataset received, it begins a processing pipeline to prepare the volume for display. Received volumes are stored and made available to users as a list of studies.
Users can also initiate loading a case on an ad-hoc basis from files already stored on the server. This can enable simpler processing of cases that are available on, for example, network attached storage, without passing through a DICOM transfer first.